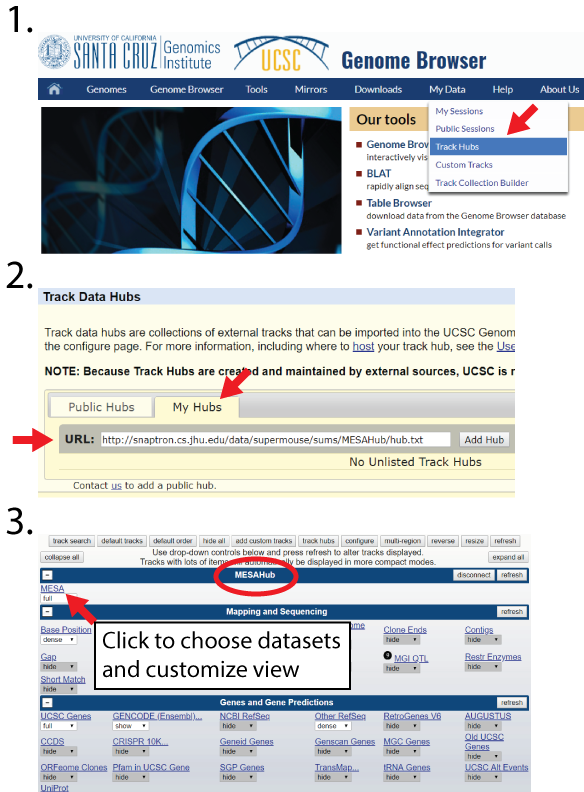
UCSC TrackHubs for data visualization:
We strongly recommend that users cross-validate any splicing results obtained from ASCOT. One way to do so is to visualize the data on the UCSC Genome Browser. We provide TrackHubs (collections of .bigwig files) from each dataset in ASCOT:
>Mouse cell types and tissues from bulk RNA-Seq (MESA) TrackHub link
http://snaptron.cs.jhu.edu/data/mesa/sums/MESAHub/hub.txt
>Mouse single-cell RNA-Seq data (CellTower) TrackHub link
http://snaptron.cs.jhu.edu/data/ct_m_s/sums/CTMSHub/hub.txt
>Human GTEx tissues + eye (GTEx) TrackHub link
http://snaptron.cs.jhu.edu/data/gtex/sums/GTEXHub/hub.txt
>ENCODE shRNA-Seq knockdown data (ENCODE) TrackHub link
http://snaptron.cs.jhu.edu/data/encode1159/sums/ENCODEHub/hub.txtInstructions for using TrackHubs are available on the UCSC help page. In brief, navigate to the top menu bar option My Data > Track Hubs, select the My Hubs tab, enter URL from above and select Add Hub.